ACRO Demonstration#
[1]:
import os
import numpy as np
import pandas as pd
[2]:
# uncomment this line if acro is not installed
# ie you are in development mode
# sys.path.insert(0, os.path.abspath(".."))
[3]:
from acro import ACRO, add_constant, add_to_acro
Instantiate ACRO#
[4]:
acro = ACRO(suppress=True)
INFO:acro:version: 0.4.8
INFO:acro:config: {'safe_threshold': 10, 'safe_dof_threshold': 10, 'safe_nk_n': 2, 'safe_nk_k': 0.9, 'safe_pratio_p': 0.1, 'check_missing_values': False, 'survival_safe_threshold': 10, 'zeros_are_disclosive': True}
INFO:acro:automatic suppression: True
Load test data#
The dataset used in this notebook is the nursery dataset from OpenML.
The dataset can be read directly from OpenML using the code commented in the next cell.
In this version, it can be read directly from the local machine if it has been downloaded.
The code below reads the data from a folder called “data” which we assume is at the same level as the folder where you are working.
The path might need to be changed if the data has been downloaded and stored elsewhere.
for example use: path = os.path.join(“data”, “nursery.arff”) if the data is in a sub-folder of your work folder
[5]:
from scipy.io.arff import loadarff
path = os.path.join("../data", "nursery.arff")
data = loadarff(path)
df = pd.DataFrame(data[0])
df = df.select_dtypes([object])
df = df.stack().str.decode("utf-8").unstack()
df.rename(columns={"class": "recommend"}, inplace=True)
df.head()
[5]:
| parents | has_nurs | form | children | housing | finance | social | health | recommend | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | usual | proper | complete | 1 | convenient | convenient | nonprob | recommended | recommend |
| 1 | usual | proper | complete | 1 | convenient | convenient | nonprob | priority | priority |
| 2 | usual | proper | complete | 1 | convenient | convenient | nonprob | not_recom | not_recom |
| 3 | usual | proper | complete | 1 | convenient | convenient | slightly_prob | recommended | recommend |
| 4 | usual | proper | complete | 1 | convenient | convenient | slightly_prob | priority | priority |
Convert ‘more than 3’ children to random between 4 and 10#
Change the children column from categorical to numeric in order to be able to test some of the ACRO functions that require a numeric feature
[6]:
print(f" children column entries in raw file {df.children.unique()}")
children column entries in raw file ['1' '2' '3' 'more']
[7]:
df["children"].replace(to_replace={"more": "4"}, inplace=True)
df["children"] = pd.to_numeric(df["children"])
df["children"] = df.apply(
lambda row: (
row["children"] if row["children"] in (1, 2, 3) else np.random.randint(4, 10)
),
axis=1,
)
Examples of producing tabular output#
first, how a researcher would normally make a call in pandas, saving the results in a variable that they can view on screen (or save to file?)
then how the call is identical in SACRO, except that:
“pd” is replaced by “acro”
the researcher immediately sees a copy of what the TRE output checker will see.
Pandas crosstab#
[8]:
table = pd.crosstab(df.recommend, df.parents)
print(table)
parents great_pret pretentious usual
recommend
not_recom 1440 1440 1440
priority 858 1484 1924
recommend 0 0 2
spec_prior 2022 1264 758
very_recom 0 132 196
ACRO crosstab#
[9]:
safe_table = acro.crosstab(
df.recommend,
df.parents,
)
print(safe_table)
INFO:acro:get_summary(): fail; threshold: 4 cells suppressed;
INFO:acro:outcome_df:
----------------------------------------------------|
parents |great_pret |pretentious |usual |
recommend | | | |
----------------------------------------------------|
not_recom | ok | ok | ok|
priority | ok | ok | ok|
recommend | threshold; | threshold; | threshold; |
spec_prior | ok | ok | ok|
very_recom | threshold; | ok | ok|
----------------------------------------------------|
INFO:acro:records:add(): output_0
parents great_pret pretentious usual
recommend
not_recom 1440.0 1440.0 1440.0
priority 858.0 1484.0 1924.0
recommend NaN NaN NaN
spec_prior 2022.0 1264.0 758.0
very_recom NaN 132.0 196.0
ACRO crosstab with totals#
[10]:
safe_table = acro.crosstab(df.recommend, df.parents, margins=True)
print(safe_table)
INFO:acro:get_summary(): fail; threshold: 5 cells suppressed;
INFO:acro:outcome_df:
------------------------------------------------------------------|
parents |great_pret |pretentious |usual |All |
recommend | | | | |
------------------------------------------------------------------|
not_recom | ok | ok | ok | ok|
priority | ok | ok | ok | ok|
recommend | threshold; | threshold; | threshold; | threshold; |
spec_prior | ok | ok | ok | ok|
very_recom | threshold; | ok | ok | ok|
All | ok | ok | ok | ok|
------------------------------------------------------------------|
INFO:acro:records:add(): output_1
parents great_pret pretentious usual All
recommend
not_recom 1440.0 1440 1440 4320
priority 858.0 1484 1924 4266
spec_prior 2022.0 1264 758 4044
very_recom NaN 132 196 328
All 4320.0 4320 4318 12958
ACRO crosstab without suppression#
[11]:
acro.suppress = False
safe_table = acro.crosstab(df.recommend, df.parents)
print(safe_table)
INFO:acro:get_summary(): fail; threshold: 4 cells may need suppressing;
INFO:acro:outcome_df:
----------------------------------------------------|
parents |great_pret |pretentious |usual |
recommend | | | |
----------------------------------------------------|
not_recom | ok | ok | ok|
priority | ok | ok | ok|
recommend | threshold; | threshold; | threshold; |
spec_prior | ok | ok | ok|
very_recom | threshold; | ok | ok|
----------------------------------------------------|
INFO:acro:records:add(): output_2
parents great_pret pretentious usual
recommend
not_recom 1440 1440 1440
priority 858 1484 1924
recommend 0 0 2
spec_prior 2022 1264 758
very_recom 0 132 196
[12]:
acro.suppress = True
ACRO crosstab with aggregation function#
[13]:
safe_table = acro.crosstab(
df.recommend, df.parents, values=df.children, aggfunc="count"
)
print(safe_table)
INFO:acro:get_summary(): fail; threshold: 1 cells suppressed; p-ratio: 4 cells suppressed; nk-rule: 4 cells suppressed;
INFO:acro:outcome_df:
------------------------------------------------------------------------------------|
parents |great_pret |pretentious |usual |
recommend | | | |
------------------------------------------------------------------------------------|
not_recom | ok | ok | ok|
priority | ok | ok | ok|
recommend | p-ratio; nk-rule; | p-ratio; nk-rule; | threshold; p-ratio; nk-rule; |
spec_prior | ok | ok | ok|
very_recom | p-ratio; nk-rule; | ok | ok|
------------------------------------------------------------------------------------|
INFO:acro:records:add(): output_3
parents great_pret pretentious usual
recommend
not_recom 1440.0 1440.0 1440.0
priority 858.0 1484.0 1924.0
recommend NaN NaN NaN
spec_prior 2022.0 1264.0 758.0
very_recom NaN 132.0 196.0
[14]:
safe_table = acro.crosstab(
df.recommend, df.parents, values=df.children, aggfunc=["mode", "mean"]
)
print(safe_table)
INFO:acro:get_summary(): fail; threshold: 2 cells suppressed; p-ratio: 8 cells suppressed; nk-rule: 8 cells suppressed;
INFO:acro:outcome_df:
---------------------------------------------------------------------------------------------------------------------------------------------------------|
mode_aggfunc |mean |
parents great_pret pretentious usual |great_pret pretentious usual |
recommend | |
---------------------------------------------------------------------------------------------------------------------------------------------------------|
not_recom ok ok ok | ok ok ok|
priority ok ok ok | ok ok ok|
recommend p-ratio; nk-rule; p-ratio; nk-rule; threshold; p-ratio; nk-rule; | p-ratio; nk-rule; p-ratio; nk-rule; threshold; p-ratio; nk-rule; |
spec_prior ok ok ok | ok ok ok|
very_recom p-ratio; nk-rule; ok ok | p-ratio; nk-rule; ok ok|
---------------------------------------------------------------------------------------------------------------------------------------------------------|
INFO:acro:records:add(): output_4
mode_aggfunc mean
parents great_pret pretentious usual great_pret pretentious usual
recommend
not_recom 2.0 1.0 1.0 3.125694 3.105556 3.074306
priority 1.0 1.0 1.0 2.665501 3.030323 3.116944
recommend NaN NaN NaN NaN NaN NaN
spec_prior 3.0 3.0 3.0 3.353610 3.370253 3.393140
very_recom NaN 1.0 1.0 NaN 2.204545 2.244898
ACRO pivot_table#
This is an example of pivot table using ACRO.
Some researchers may prefer this to using crosstab.
Again the call syntax is identical to the pandas “pd.pivot_table”
in this case the output is non-disclosive
[15]:
table = acro.pivot_table(
df, index=["parents"], values=["children"], aggfunc=["mean", "std"]
)
print(table)
INFO:acro:get_summary(): pass
INFO:acro:outcome_df:
------------------------------|
mean |std |
children |children|
parents | |
------------------------------|
great_pret ok | ok |
pretentious ok | ok |
usual ok | ok |
------------------------------|
INFO:acro:records:add(): output_5
mean std
children children
parents
great_pret 3.140972 2.270396
pretentious 3.129630 2.250436
usual 3.110648 2.213072
ACRO pivot_table with margins#
[16]:
safe_table = acro.pivot_table(
df, columns=["recommend"], index=["parents"], values=["children"], margins=True
)
print(safe_table)
INFO:acro:get_summary(): fail; threshold: 5 cells suppressed; p-ratio: 5 cells suppressed; nk-rule: 5 cells suppressed;
INFO:acro:outcome_df:
-----------------------------------------------------------------------------------------------------------|
children |
recommend not_recom priority recommend spec_prior very_recom All|
parents |
-----------------------------------------------------------------------------------------------------------|
great_pret ok ok threshold; p-ratio; nk-rule; ok threshold; p-ratio; nk-rule; ok|
pretentious ok ok threshold; p-ratio; nk-rule; ok ok ok|
usual ok ok threshold; p-ratio; nk-rule; ok ok ok|
All ok ok threshold; p-ratio; nk-rule; ok ok ok|
-----------------------------------------------------------------------------------------------------------|
INFO:acro:Disclosive cells were deleted from the dataframe before calculating the pivot table
INFO:acro:records:add(): output_6
children
recommend not_recom priority spec_prior very_recom All
parents
great_pret 3.125694 2.665501 3.353610 NaN 3.140972
pretentious 3.105556 3.030323 3.370253 2.204545 3.129630
usual 3.074306 3.116944 3.393140 2.244898 3.111626
All 3.101852 2.996015 3.366222 2.228659 3.127412
Regression examples using ACRO#
Again there is an industry-standard package in python, this time called statsmodels.
The examples below illustrate the use of the ACRO wrapper standard statsmodel functions
Note that statsmodels can be called using an ‘R-like’ format (using an ‘r’ suffix on the command names)
most statsmodels functiobns return a “results object” which has a “summary” function that produces printable/saveable outputs
Start by manipulating the nursery data to get two numeric variables#
The ‘recommend’ column is converted to an integer scale
[17]:
df["recommend"].replace(
to_replace={
"not_recom": "0",
"recommend": "1",
"very_recom": "2",
"priority": "3",
"spec_prior": "4",
},
inplace=True,
)
df["recommend"] = pd.to_numeric(df["recommend"])
new_df = df[["recommend", "children"]]
new_df = new_df.dropna()
ACRO OLS#
This is an example of ordinary least square regression using ACRO.
Above recommend column was converted form categorical to numeric.
Now we perform a the linear regression between recommend and children.
This version includes a constant (intercept)
This is just to show how the regression is done using ACRO.
No correlation is expected to be seen by using these variables
[18]:
y = new_df["recommend"]
x = new_df["children"]
x = add_constant(x)
results = acro.ols(y, x)
results.summary()
INFO:acro:ols() outcome: pass; dof=12958.0 >= 10
INFO:acro:records:add(): output_7
[18]:
| Dep. Variable: | recommend | R-squared: | 0.001 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.001 |
| Method: | Least Squares | F-statistic: | 13.83 |
| Date: | Thu, 06 Mar 2025 | Prob (F-statistic): | 0.000201 |
| Time: | 19:39:47 | Log-Likelihood: | -25121. |
| No. Observations: | 12960 | AIC: | 5.025e+04 |
| Df Residuals: | 12958 | BIC: | 5.026e+04 |
| Df Model: | 1 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| const | 2.2099 | 0.025 | 87.263 | 0.000 | 2.160 | 2.260 |
| children | 0.0245 | 0.007 | 3.718 | 0.000 | 0.012 | 0.037 |
| Omnibus: | 77090.215 | Durbin-Watson: | 2.883 |
|---|---|---|---|
| Prob(Omnibus): | 0.000 | Jarque-Bera (JB): | 1741.570 |
| Skew: | -0.486 | Prob(JB): | 0.00 |
| Kurtosis: | 1.489 | Cond. No. | 6.90 |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
[18]:
| Dep. Variable: | recommend | R-squared: | 0.000 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.000 |
| Method: | Least Squares | F-statistic: | 6.417 |
| Date: | Mon, 04 Mar 2024 | Prob (F-statistic): | 0.0113 |
| Time: | 21:21:09 | Log-Likelihood: | -25124. |
| No. Observations: | 12960 | AIC: | 5.025e+04 |
| Df Residuals: | 12958 | BIC: | 5.027e+04 |
| Df Model: | 1 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| const | 2.2341 | 0.025 | 87.965 | 0.000 | 2.184 | 2.284 |
| children | 0.0168 | 0.007 | 2.533 | 0.011 | 0.004 | 0.030 |
| Omnibus: | 76735.931 | Durbin-Watson: | 2.883 |
|---|---|---|---|
| Prob(Omnibus): | 0.000 | Jarque-Bera (JB): | 1742.843 |
| Skew: | -0.485 | Prob(JB): | 0.00 |
| Kurtosis: | 1.487 | Cond. No. | 6.89 |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
ACRO OLSR#
This is an example of ordinary least squares regression using the ‘R-like’ statsmodels api, i.e. from a formula and dataframe using ACRO
[19]:
results = acro.olsr(formula="recommend ~ children", data=new_df)
print(results.summary())
INFO:acro:olsr() outcome: pass; dof=12958.0 >= 10
INFO:acro:records:add(): output_8
OLS Regression Results
==============================================================================
Dep. Variable: recommend R-squared: 0.001
Model: OLS Adj. R-squared: 0.001
Method: Least Squares F-statistic: 13.83
Date: Thu, 06 Mar 2025 Prob (F-statistic): 0.000201
Time: 19:39:47 Log-Likelihood: -25121.
No. Observations: 12960 AIC: 5.025e+04
Df Residuals: 12958 BIC: 5.026e+04
Df Model: 1
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
Intercept 2.2099 0.025 87.263 0.000 2.160 2.260
children 0.0245 0.007 3.718 0.000 0.012 0.037
==============================================================================
Omnibus: 77090.215 Durbin-Watson: 2.883
Prob(Omnibus): 0.000 Jarque-Bera (JB): 1741.570
Skew: -0.486 Prob(JB): 0.00
Kurtosis: 1.489 Cond. No. 6.90
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
OLS Regression Results
==============================================================================
Dep. Variable: recommend R-squared: 0.000
Model: OLS Adj. R-squared: 0.000
Method: Least Squares F-statistic: 6.417
Date: Mon, 04 Mar 2024 Prob (F-statistic): 0.0113
Time: 21:21:09 Log-Likelihood: -25124.
No. Observations: 12960 AIC: 5.025e+04
Df Residuals: 12958 BIC: 5.027e+04
Df Model: 1
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
Intercept 2.2341 0.025 87.965 0.000 2.184 2.284
children 0.0168 0.007 2.533 0.011 0.004 0.030
==============================================================================
Omnibus: 76735.931 Durbin-Watson: 2.883
Prob(Omnibus): 0.000 Jarque-Bera (JB): 1742.843
Skew: -0.485 Prob(JB): 0.00
Kurtosis: 1.487 Cond. No. 6.89
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
ACRO Probit#
Again, we support the use of R-like formulas - because we support R
[20]:
new_df = df[["finance", "children"]]
new_df = new_df.dropna()
y = new_df["finance"].astype("category").cat.codes # numeric
y.name = "finance"
x = new_df["children"]
x = add_constant(x)
results = acro.probit(y, x)
print(results.summary())
INFO:acro:probit() outcome: pass; dof=12958.0 >= 10
Optimization terminated successfully.
Current function value: 0.693145
Iterations 2
INFO:acro:records:add(): output_9
Probit Regression Results
==============================================================================
Dep. Variable: finance No. Observations: 12960
Model: Probit Df Residuals: 12958
Method: MLE Df Model: 1
Date: Thu, 06 Mar 2025 Pseudo R-squ.: 3.602e-06
Time: 19:39:47 Log-Likelihood: -8983.2
converged: True LL-Null: -8983.2
Covariance Type: nonrobust LLR p-value: 0.7992
==============================================================================
coef std err z P>|z| [0.025 0.975]
------------------------------------------------------------------------------
const -0.0039 0.019 -0.207 0.836 -0.041 0.033
children 0.0012 0.005 0.254 0.799 -0.008 0.011
==============================================================================
ACRO Logit#
This is an example of logistic regression using ACRO using the statmodels function
[21]:
results = acro.logit(y, x)
results.summary()
INFO:acro:logit() outcome: pass; dof=12958.0 >= 10
INFO:acro:records:add(): output_10
Optimization terminated successfully.
Current function value: 0.693145
Iterations 3
[21]:
| Dep. Variable: | finance | No. Observations: | 12960 |
|---|---|---|---|
| Model: | Logit | Df Residuals: | 12958 |
| Method: | MLE | Df Model: | 1 |
| Date: | Thu, 06 Mar 2025 | Pseudo R-squ.: | 3.602e-06 |
| Time: | 19:39:47 | Log-Likelihood: | -8983.2 |
| converged: | True | LL-Null: | -8983.2 |
| Covariance Type: | nonrobust | LLR p-value: | 0.7992 |
| coef | std err | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| const | -0.0062 | 0.030 | -0.207 | 0.836 | -0.065 | 0.053 |
| children | 0.0020 | 0.008 | 0.254 | 0.799 | -0.013 | 0.017 |
[21]:
| Dep. Variable: | finance | No. Observations: | 12960 |
|---|---|---|---|
| Model: | Logit | Df Residuals: | 12958 |
| Method: | MLE | Df Model: | 1 |
| Date: | Mon, 04 Mar 2024 | Pseudo R-squ.: | 1.186e-06 |
| Time: | 21:21:09 | Log-Likelihood: | -8983.2 |
| converged: | True | LL-Null: | -8983.2 |
| Covariance Type: | nonrobust | LLR p-value: | 0.8839 |
| coef | std err | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| const | -0.0036 | 0.030 | -0.119 | 0.905 | -0.063 | 0.056 |
| children | 0.0012 | 0.008 | 0.146 | 0.884 | -0.014 | 0.017 |
ACRO survival analysis#
This is an example of survival tables and plots using ACRO.
A dataset from statsmodels is used for the survival analysis.
A subset of tha dataset is used in this example to demostrate the survival analysis.
The output parameter in the surv_func define the type of output (table or plot).
[22]:
import statsmodels.api as sm
data = sm.datasets.get_rdataset("flchain", "survival").data
data = data.loc[data.sex == "F", :]
data = data.iloc[:20, :]
# data.head()
[23]:
safe_table = acro.surv_func(data.futime, data.death, output="table")
print(safe_table)
INFO:acro:get_summary(): fail; threshold: 76 cells suppressed;
INFO:acro:outcome_df:
-----------------------------------------------------------|
Surv_prob |Surv_prob_SE |num_at_risk |num_events |
Time | | | |
-----------------------------------------------------------|
51 ok | ok | ok | ok|
69 threshold; | threshold; | threshold; | threshold; |
85 threshold; | threshold; | threshold; | threshold; |
91 threshold; | threshold; | threshold; | threshold; |
115 threshold; | threshold; | threshold; | threshold; |
372 threshold; | threshold; | threshold; | threshold; |
667 threshold; | threshold; | threshold; | threshold; |
874 threshold; | threshold; | threshold; | threshold; |
1039 threshold; | threshold; | threshold; | threshold; |
1046 threshold; | threshold; | threshold; | threshold; |
1281 threshold; | threshold; | threshold; | threshold; |
1286 threshold; | threshold; | threshold; | threshold; |
1326 threshold; | threshold; | threshold; | threshold; |
1355 threshold; | threshold; | threshold; | threshold; |
1626 threshold; | threshold; | threshold; | threshold; |
1903 threshold; | threshold; | threshold; | threshold; |
1914 threshold; | threshold; | threshold; | threshold; |
2776 threshold; | threshold; | threshold; | threshold; |
2851 threshold; | threshold; | threshold; | threshold; |
3309 threshold; | threshold; | threshold; | threshold; |
-----------------------------------------------------------|
INFO:acro:records:add(): output_11
Surv prob Surv prob SE num at risk num events
Time
51 0.95 0.048734 20.0 1.0
69 NaN NaN NaN NaN
85 NaN NaN NaN NaN
91 NaN NaN NaN NaN
115 NaN NaN NaN NaN
372 NaN NaN NaN NaN
667 NaN NaN NaN NaN
874 NaN NaN NaN NaN
1039 NaN NaN NaN NaN
1046 NaN NaN NaN NaN
1281 NaN NaN NaN NaN
1286 NaN NaN NaN NaN
1326 NaN NaN NaN NaN
1355 NaN NaN NaN NaN
1626 NaN NaN NaN NaN
1903 NaN NaN NaN NaN
1914 NaN NaN NaN NaN
2776 NaN NaN NaN NaN
2851 NaN NaN NaN NaN
3309 NaN NaN NaN NaN
[24]:
safe_plot = acro.surv_func(
data.futime, data.death, output="plot", filename="kaplan-mier.png"
)
print(safe_plot)
INFO:acro:get_summary(): fail; threshold: 76 cells suppressed;
INFO:acro:outcome_df:
-----------------------------------------------------------|
Surv_prob |Surv_prob_SE |num_at_risk |num_events |
Time | | | |
-----------------------------------------------------------|
51 ok | ok | ok | ok|
69 threshold; | threshold; | threshold; | threshold; |
85 threshold; | threshold; | threshold; | threshold; |
91 threshold; | threshold; | threshold; | threshold; |
115 threshold; | threshold; | threshold; | threshold; |
372 threshold; | threshold; | threshold; | threshold; |
667 threshold; | threshold; | threshold; | threshold; |
874 threshold; | threshold; | threshold; | threshold; |
1039 threshold; | threshold; | threshold; | threshold; |
1046 threshold; | threshold; | threshold; | threshold; |
1281 threshold; | threshold; | threshold; | threshold; |
1286 threshold; | threshold; | threshold; | threshold; |
1326 threshold; | threshold; | threshold; | threshold; |
1355 threshold; | threshold; | threshold; | threshold; |
1626 threshold; | threshold; | threshold; | threshold; |
1903 threshold; | threshold; | threshold; | threshold; |
1914 threshold; | threshold; | threshold; | threshold; |
2776 threshold; | threshold; | threshold; | threshold; |
2851 threshold; | threshold; | threshold; | threshold; |
3309 threshold; | threshold; | threshold; | threshold; |
-----------------------------------------------------------|
INFO:acro:records:add(): output_12
(<Axes: xlabel='Time'>, 'acro_artifacts/kaplan-mier_0.png')
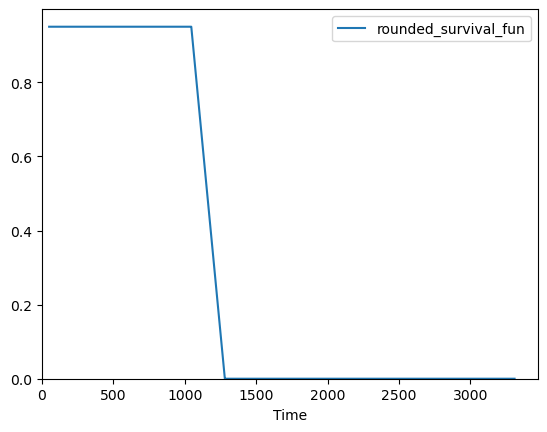
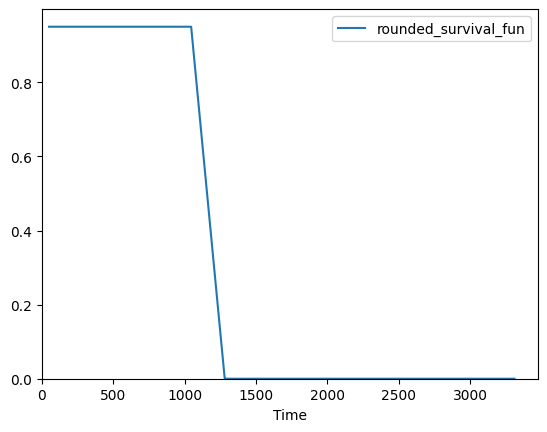
ACRO functionality to let users manage their outputs#
1: List current ACRO outputs#
This is an example of using the print_output function to list all the outputs created so far
[25]:
acro.print_outputs()
uid: output_0
status: fail
type: table
properties: {'method': 'crosstab'}
sdc: {'summary': {'suppressed': True, 'negative': 0, 'missing': 0, 'threshold': 4, 'p-ratio': 0, 'nk-rule': 0, 'all-values-are-same': 0}, 'cells': {'negative': [], 'missing': [], 'threshold': [[2, 0], [2, 1], [2, 2], [4, 0]], 'p-ratio': [], 'nk-rule': [], 'all-values-are-same': []}}
command: safe_table = acro.crosstab(
summary: fail; threshold: 4 cells suppressed;
outcome: parents great_pret pretentious usual
recommend
not_recom ok ok ok
priority ok ok ok
recommend threshold; threshold; threshold;
spec_prior ok ok ok
very_recom threshold; ok ok
output: [parents great_pret pretentious usual
recommend
not_recom 1440.0 1440.0 1440.0
priority 858.0 1484.0 1924.0
recommend NaN NaN NaN
spec_prior 2022.0 1264.0 758.0
very_recom NaN 132.0 196.0]
timestamp: 2025-03-06T19:39:46.897407
comments: []
exception:
uid: output_1
status: fail
type: table
properties: {'method': 'crosstab'}
sdc: {'summary': {'suppressed': True, 'negative': 0, 'missing': 0, 'threshold': 5, 'p-ratio': 0, 'nk-rule': 0, 'all-values-are-same': 0}, 'cells': {'negative': [], 'missing': [], 'threshold': [[2, 0], [2, 1], [2, 2], [2, 3], [4, 0]], 'p-ratio': [], 'nk-rule': [], 'all-values-are-same': []}}
command: safe_table = acro.crosstab(df.recommend, df.parents, margins=True)
summary: fail; threshold: 5 cells suppressed;
outcome: parents great_pret pretentious usual All
recommend
not_recom ok ok ok ok
priority ok ok ok ok
recommend threshold; threshold; threshold; threshold;
spec_prior ok ok ok ok
very_recom threshold; ok ok ok
All ok ok ok ok
output: [parents great_pret pretentious usual All
recommend
not_recom 1440.0 1440 1440 4320
priority 858.0 1484 1924 4266
spec_prior 2022.0 1264 758 4044
very_recom NaN 132 196 328
All 4320.0 4320 4318 12958]
timestamp: 2025-03-06T19:39:46.961631
comments: []
exception:
uid: output_2
status: fail
type: table
properties: {'method': 'crosstab'}
sdc: {'summary': {'suppressed': False, 'negative': 0, 'missing': 0, 'threshold': 4, 'p-ratio': 0, 'nk-rule': 0, 'all-values-are-same': 0}, 'cells': {'negative': [], 'missing': [], 'threshold': [[2, 0], [2, 1], [2, 2], [4, 0]], 'p-ratio': [], 'nk-rule': [], 'all-values-are-same': []}}
command: safe_table = acro.crosstab(df.recommend, df.parents)
summary: fail; threshold: 4 cells may need suppressing;
outcome: parents great_pret pretentious usual
recommend
not_recom ok ok ok
priority ok ok ok
recommend threshold; threshold; threshold;
spec_prior ok ok ok
very_recom threshold; ok ok
output: [parents great_pret pretentious usual
recommend
not_recom 1440 1440 1440
priority 858 1484 1924
recommend 0 0 2
spec_prior 2022 1264 758
very_recom 0 132 196]
timestamp: 2025-03-06T19:39:46.980090
comments: []
exception:
uid: output_3
status: fail
type: table
properties: {'method': 'crosstab'}
sdc: {'summary': {'suppressed': True, 'negative': 0, 'missing': 0, 'threshold': 1, 'p-ratio': 4, 'nk-rule': 4, 'all-values-are-same': 0}, 'cells': {'negative': [], 'missing': [], 'threshold': [[2, 2]], 'p-ratio': [[2, 0], [2, 1], [2, 2], [4, 0]], 'nk-rule': [[2, 0], [2, 1], [2, 2], [4, 0]], 'all-values-are-same': []}}
command: safe_table = acro.crosstab(
summary: fail; threshold: 1 cells suppressed; p-ratio: 4 cells suppressed; nk-rule: 4 cells suppressed;
outcome: parents great_pret pretentious \
recommend
not_recom ok ok
priority ok ok
recommend p-ratio; nk-rule; p-ratio; nk-rule;
spec_prior ok ok
very_recom p-ratio; nk-rule; ok
parents usual
recommend
not_recom ok
priority ok
recommend threshold; p-ratio; nk-rule;
spec_prior ok
very_recom ok
output: [parents great_pret pretentious usual
recommend
not_recom 1440.0 1440.0 1440.0
priority 858.0 1484.0 1924.0
recommend NaN NaN NaN
spec_prior 2022.0 1264.0 758.0
very_recom NaN 132.0 196.0]
timestamp: 2025-03-06T19:39:47.019919
comments: []
exception:
uid: output_4
status: fail
type: table
properties: {'method': 'crosstab'}
sdc: {'summary': {'suppressed': True, 'negative': 0, 'missing': 0, 'threshold': 2, 'p-ratio': 8, 'nk-rule': 8, 'all-values-are-same': 0}, 'cells': {'negative': [], 'missing': [], 'threshold': [[2, 2], [2, 5]], 'p-ratio': [[2, 0], [2, 1], [2, 2], [2, 3], [2, 4], [2, 5], [4, 0], [4, 3]], 'nk-rule': [[2, 0], [2, 1], [2, 2], [2, 3], [2, 4], [2, 5], [4, 0], [4, 3]], 'all-values-are-same': []}}
command: safe_table = acro.crosstab(
summary: fail; threshold: 2 cells suppressed; p-ratio: 8 cells suppressed; nk-rule: 8 cells suppressed;
outcome: mode_aggfunc \
parents great_pret pretentious
recommend
not_recom ok ok
priority ok ok
recommend p-ratio; nk-rule; p-ratio; nk-rule;
spec_prior ok ok
very_recom p-ratio; nk-rule; ok
mean \
parents usual great_pret
recommend
not_recom ok ok
priority ok ok
recommend threshold; p-ratio; nk-rule; p-ratio; nk-rule;
spec_prior ok ok
very_recom ok p-ratio; nk-rule;
parents pretentious usual
recommend
not_recom ok ok
priority ok ok
recommend p-ratio; nk-rule; threshold; p-ratio; nk-rule;
spec_prior ok ok
very_recom ok ok
output: [ mode_aggfunc mean
parents great_pret pretentious usual great_pret pretentious usual
recommend
not_recom 2.0 1.0 1.0 3.125694 3.105556 3.074306
priority 1.0 1.0 1.0 2.665501 3.030323 3.116944
recommend NaN NaN NaN NaN NaN NaN
spec_prior 3.0 3.0 3.0 3.353610 3.370253 3.393140
very_recom NaN 1.0 1.0 NaN 2.204545 2.244898]
timestamp: 2025-03-06T19:39:47.068066
comments: []
exception:
uid: output_5
status: pass
type: table
properties: {'method': 'pivot_table'}
sdc: {'summary': {'suppressed': True, 'negative': 0, 'missing': 0, 'threshold': 0, 'p-ratio': 0, 'nk-rule': 0, 'all-values-are-same': 0}, 'cells': {'negative': [], 'missing': [], 'threshold': [], 'p-ratio': [], 'nk-rule': [], 'all-values-are-same': []}}
command: table = acro.pivot_table(
summary: pass
outcome: mean std
children children
parents
great_pret ok ok
pretentious ok ok
usual ok ok
output: [ mean std
children children
parents
great_pret 3.140972 2.270396
pretentious 3.129630 2.250436
usual 3.110648 2.213072]
timestamp: 2025-03-06T19:39:47.105651
comments: []
exception:
uid: output_6
status: fail
type: table
properties: {'method': 'pivot_table'}
sdc: {'summary': {'suppressed': True, 'negative': 0, 'missing': 0, 'threshold': 5, 'p-ratio': 5, 'nk-rule': 5, 'all-values-are-same': 0}, 'cells': {'negative': [], 'missing': [], 'threshold': [[0, 2], [0, 4], [1, 2], [2, 2], [3, 2]], 'p-ratio': [[0, 2], [0, 4], [1, 2], [2, 2], [3, 2]], 'nk-rule': [[0, 2], [0, 4], [1, 2], [2, 2], [3, 2]], 'all-values-are-same': []}}
command: safe_table = acro.pivot_table(
summary: fail; threshold: 5 cells suppressed; p-ratio: 5 cells suppressed; nk-rule: 5 cells suppressed;
outcome: children \
recommend not_recom priority recommend spec_prior
parents
great_pret ok ok threshold; p-ratio; nk-rule; ok
pretentious ok ok threshold; p-ratio; nk-rule; ok
usual ok ok threshold; p-ratio; nk-rule; ok
All ok ok threshold; p-ratio; nk-rule; ok
recommend very_recom All
parents
great_pret threshold; p-ratio; nk-rule; ok
pretentious ok ok
usual ok ok
All ok ok
output: [ children
recommend not_recom priority spec_prior very_recom All
parents
great_pret 3.125694 2.665501 3.353610 NaN 3.140972
pretentious 3.105556 3.030323 3.370253 2.204545 3.129630
usual 3.074306 3.116944 3.393140 2.244898 3.111626
All 3.101852 2.996015 3.366222 2.228659 3.127412]
timestamp: 2025-03-06T19:39:47.231513
comments: []
exception:
uid: output_7
status: pass
type: regression
properties: {'method': 'ols', 'dof': 12958.0}
sdc: {}
command: results = acro.ols(y, x)
summary: pass; dof=12958.0 >= 10
outcome: Empty DataFrame
Columns: []
Index: []
output: [ recommend R-squared: 0.001
Dep. Variable:
Model: OLS Adj. R-squared: 0.001000
Method: Least Squares F-statistic: 13.830000
Date: Thu, 06 Mar 2025 Prob (F-statistic): 0.000201
Time: 19:39:47 Log-Likelihood: -25121.000000
No. Observations: 12960 AIC: 50250.000000
Df Residuals: 12958 BIC: 50260.000000
Df Model: 1 NaN NaN
Covariance Type: nonrobust NaN NaN, coef std err t P>|t| [0.025 0.975]
const 2.2099 0.025 87.263 0.0 2.160 2.260
children 0.0245 0.007 3.718 0.0 0.012 0.037, 77090.215 Durbin-Watson: 2.883
Omnibus:
Prob(Omnibus): 0.000 Jarque-Bera (JB): 1741.57
Skew: -0.486 Prob(JB): 0.00
Kurtosis: 1.489 Cond. No. 6.90]
timestamp: 2025-03-06T19:39:47.388052
comments: []
exception:
uid: output_8
status: pass
type: regression
properties: {'method': 'olsr', 'dof': 12958.0}
sdc: {}
command: results = acro.olsr(formula="recommend ~ children", data=new_df)
summary: pass; dof=12958.0 >= 10
outcome: Empty DataFrame
Columns: []
Index: []
output: [ recommend R-squared: 0.001
Dep. Variable:
Model: OLS Adj. R-squared: 0.001000
Method: Least Squares F-statistic: 13.830000
Date: Thu, 06 Mar 2025 Prob (F-statistic): 0.000201
Time: 19:39:47 Log-Likelihood: -25121.000000
No. Observations: 12960 AIC: 50250.000000
Df Residuals: 12958 BIC: 50260.000000
Df Model: 1 NaN NaN
Covariance Type: nonrobust NaN NaN, coef std err t P>|t| [0.025 0.975]
Intercept 2.2099 0.025 87.263 0.0 2.160 2.260
children 0.0245 0.007 3.718 0.0 0.012 0.037, 77090.215 Durbin-Watson: 2.883
Omnibus:
Prob(Omnibus): 0.000 Jarque-Bera (JB): 1741.57
Skew: -0.486 Prob(JB): 0.00
Kurtosis: 1.489 Cond. No. 6.90]
timestamp: 2025-03-06T19:39:47.414293
comments: []
exception:
uid: output_9
status: pass
type: regression
properties: {'method': 'probit', 'dof': 12958.0}
sdc: {}
command: results = acro.probit(y, x)
summary: pass; dof=12958.0 >= 10
outcome: Empty DataFrame
Columns: []
Index: []
output: [ finance No. Observations: 12960
Dep. Variable:
Model: Probit Df Residuals: 12958.000000
Method: MLE Df Model: 1.000000
Date: Thu, 06 Mar 2025 Pseudo R-squ.: 0.000004
Time: 19:39:47 Log-Likelihood: -8983.200000
converged: True LL-Null: -8983.200000
Covariance Type: nonrobust LLR p-value: 0.799200, coef std err z P>|z| [0.025 0.975]
const -0.0039 0.019 -0.207 0.836 -0.041 0.033
children 0.0012 0.005 0.254 0.799 -0.008 0.011]
timestamp: 2025-03-06T19:39:47.439598
comments: []
exception:
uid: output_10
status: pass
type: regression
properties: {'method': 'logit', 'dof': 12958.0}
sdc: {}
command: results = acro.logit(y, x)
summary: pass; dof=12958.0 >= 10
outcome: Empty DataFrame
Columns: []
Index: []
output: [ finance No. Observations: 12960
Dep. Variable:
Model: Logit Df Residuals: 12958.000000
Method: MLE Df Model: 1.000000
Date: Thu, 06 Mar 2025 Pseudo R-squ.: 0.000004
Time: 19:39:47 Log-Likelihood: -8983.200000
converged: True LL-Null: -8983.200000
Covariance Type: nonrobust LLR p-value: 0.799200, coef std err z P>|z| [0.025 0.975]
const -0.0062 0.030 -0.207 0.836 -0.065 0.053
children 0.0020 0.008 0.254 0.799 -0.013 0.017]
timestamp: 2025-03-06T19:39:47.457696
comments: []
exception:
uid: output_11
status: fail
type: table
properties: {'method': 'surv_func'}
sdc: {'summary': {'suppressed': True, 'negative': 0, 'missing': 0, 'threshold': 76, 'p-ratio': 0, 'nk-rule': 0, 'all-values-are-same': 0}, 'cells': {'negative': [], 'missing': [], 'threshold': [[1, 0], [1, 1], [1, 2], [1, 3], [2, 0], [2, 1], [2, 2], [2, 3], [3, 0], [3, 1], [3, 2], [3, 3], [4, 0], [4, 1], [4, 2], [4, 3], [5, 0], [5, 1], [5, 2], [5, 3], [6, 0], [6, 1], [6, 2], [6, 3], [7, 0], [7, 1], [7, 2], [7, 3], [8, 0], [8, 1], [8, 2], [8, 3], [9, 0], [9, 1], [9, 2], [9, 3], [10, 0], [10, 1], [10, 2], [10, 3], [11, 0], [11, 1], [11, 2], [11, 3], [12, 0], [12, 1], [12, 2], [12, 3], [13, 0], [13, 1], [13, 2], [13, 3], [14, 0], [14, 1], [14, 2], [14, 3], [15, 0], [15, 1], [15, 2], [15, 3], [16, 0], [16, 1], [16, 2], [16, 3], [17, 0], [17, 1], [17, 2], [17, 3], [18, 0], [18, 1], [18, 2], [18, 3], [19, 0], [19, 1], [19, 2], [19, 3]], 'p-ratio': [], 'nk-rule': [], 'all-values-are-same': []}}
command: safe_table = acro.surv_func(data.futime, data.death, output="table")
summary: fail; threshold: 76 cells suppressed;
outcome: Surv_prob Surv_prob_SE num_at_risk num_events
Time
51 ok ok ok ok
69 threshold; threshold; threshold; threshold;
85 threshold; threshold; threshold; threshold;
91 threshold; threshold; threshold; threshold;
115 threshold; threshold; threshold; threshold;
372 threshold; threshold; threshold; threshold;
667 threshold; threshold; threshold; threshold;
874 threshold; threshold; threshold; threshold;
1039 threshold; threshold; threshold; threshold;
1046 threshold; threshold; threshold; threshold;
1281 threshold; threshold; threshold; threshold;
1286 threshold; threshold; threshold; threshold;
1326 threshold; threshold; threshold; threshold;
1355 threshold; threshold; threshold; threshold;
1626 threshold; threshold; threshold; threshold;
1903 threshold; threshold; threshold; threshold;
1914 threshold; threshold; threshold; threshold;
2776 threshold; threshold; threshold; threshold;
2851 threshold; threshold; threshold; threshold;
3309 threshold; threshold; threshold; threshold;
output: [ Surv prob Surv prob SE num at risk num events
Time
51 0.95 0.048734 20.0 1.0
69 NaN NaN NaN NaN
85 NaN NaN NaN NaN
91 NaN NaN NaN NaN
115 NaN NaN NaN NaN
372 NaN NaN NaN NaN
667 NaN NaN NaN NaN
874 NaN NaN NaN NaN
1039 NaN NaN NaN NaN
1046 NaN NaN NaN NaN
1281 NaN NaN NaN NaN
1286 NaN NaN NaN NaN
1326 NaN NaN NaN NaN
1355 NaN NaN NaN NaN
1626 NaN NaN NaN NaN
1903 NaN NaN NaN NaN
1914 NaN NaN NaN NaN
2776 NaN NaN NaN NaN
2851 NaN NaN NaN NaN
3309 NaN NaN NaN NaN]
timestamp: 2025-03-06T19:39:48.298262
comments: []
exception:
uid: output_12
status: fail
type: survival plot
properties: {'method': 'surv_func'}
sdc: {'summary': {'suppressed': True, 'negative': 0, 'missing': 0, 'threshold': 76, 'p-ratio': 0, 'nk-rule': 0, 'all-values-are-same': 0}, 'cells': {'negative': [], 'missing': [], 'threshold': [[1, 0], [1, 1], [1, 2], [1, 3], [2, 0], [2, 1], [2, 2], [2, 3], [3, 0], [3, 1], [3, 2], [3, 3], [4, 0], [4, 1], [4, 2], [4, 3], [5, 0], [5, 1], [5, 2], [5, 3], [6, 0], [6, 1], [6, 2], [6, 3], [7, 0], [7, 1], [7, 2], [7, 3], [8, 0], [8, 1], [8, 2], [8, 3], [9, 0], [9, 1], [9, 2], [9, 3], [10, 0], [10, 1], [10, 2], [10, 3], [11, 0], [11, 1], [11, 2], [11, 3], [12, 0], [12, 1], [12, 2], [12, 3], [13, 0], [13, 1], [13, 2], [13, 3], [14, 0], [14, 1], [14, 2], [14, 3], [15, 0], [15, 1], [15, 2], [15, 3], [16, 0], [16, 1], [16, 2], [16, 3], [17, 0], [17, 1], [17, 2], [17, 3], [18, 0], [18, 1], [18, 2], [18, 3], [19, 0], [19, 1], [19, 2], [19, 3]], 'p-ratio': [], 'nk-rule': [], 'all-values-are-same': []}}
command: safe_plot = acro.surv_func(
summary: fail; threshold: 76 cells suppressed;
outcome: Empty DataFrame
Columns: []
Index: []
output: ['acro_artifacts/kaplan-mier_0.png']
timestamp: 2025-03-06T19:39:48.450221
comments: []
exception:
[25]:
'uid: output_0\nstatus: fail\ntype: table\nproperties: {\'method\': \'crosstab\'}\nsdc: {\'summary\': {\'suppressed\': True, \'negative\': 0, \'missing\': 0, \'threshold\': 4, \'p-ratio\': 0, \'nk-rule\': 0, \'all-values-are-same\': 0}, \'cells\': {\'negative\': [], \'missing\': [], \'threshold\': [[2, 0], [2, 1], [2, 2], [4, 0]], \'p-ratio\': [], \'nk-rule\': [], \'all-values-are-same\': []}}\ncommand: safe_table = acro.crosstab(\nsummary: fail; threshold: 4 cells suppressed; \noutcome: parents great_pret pretentious usual\nrecommend \nnot_recom ok ok ok\npriority ok ok ok\nrecommend threshold; threshold; threshold; \nspec_prior ok ok ok\nvery_recom threshold; ok ok\noutput: [parents great_pret pretentious usual\nrecommend \nnot_recom 1440.0 1440.0 1440.0\npriority 858.0 1484.0 1924.0\nrecommend NaN NaN NaN\nspec_prior 2022.0 1264.0 758.0\nvery_recom NaN 132.0 196.0]\ntimestamp: 2025-03-06T19:39:46.897407\ncomments: []\nexception: \n\nuid: output_1\nstatus: fail\ntype: table\nproperties: {\'method\': \'crosstab\'}\nsdc: {\'summary\': {\'suppressed\': True, \'negative\': 0, \'missing\': 0, \'threshold\': 5, \'p-ratio\': 0, \'nk-rule\': 0, \'all-values-are-same\': 0}, \'cells\': {\'negative\': [], \'missing\': [], \'threshold\': [[2, 0], [2, 1], [2, 2], [2, 3], [4, 0]], \'p-ratio\': [], \'nk-rule\': [], \'all-values-are-same\': []}}\ncommand: safe_table = acro.crosstab(df.recommend, df.parents, margins=True)\nsummary: fail; threshold: 5 cells suppressed; \noutcome: parents great_pret pretentious usual All\nrecommend \nnot_recom ok ok ok ok\npriority ok ok ok ok\nrecommend threshold; threshold; threshold; threshold; \nspec_prior ok ok ok ok\nvery_recom threshold; ok ok ok\nAll ok ok ok ok\noutput: [parents great_pret pretentious usual All\nrecommend \nnot_recom 1440.0 1440 1440 4320\npriority 858.0 1484 1924 4266\nspec_prior 2022.0 1264 758 4044\nvery_recom NaN 132 196 328\nAll 4320.0 4320 4318 12958]\ntimestamp: 2025-03-06T19:39:46.961631\ncomments: []\nexception: \n\nuid: output_2\nstatus: fail\ntype: table\nproperties: {\'method\': \'crosstab\'}\nsdc: {\'summary\': {\'suppressed\': False, \'negative\': 0, \'missing\': 0, \'threshold\': 4, \'p-ratio\': 0, \'nk-rule\': 0, \'all-values-are-same\': 0}, \'cells\': {\'negative\': [], \'missing\': [], \'threshold\': [[2, 0], [2, 1], [2, 2], [4, 0]], \'p-ratio\': [], \'nk-rule\': [], \'all-values-are-same\': []}}\ncommand: safe_table = acro.crosstab(df.recommend, df.parents)\nsummary: fail; threshold: 4 cells may need suppressing; \noutcome: parents great_pret pretentious usual\nrecommend \nnot_recom ok ok ok\npriority ok ok ok\nrecommend threshold; threshold; threshold; \nspec_prior ok ok ok\nvery_recom threshold; ok ok\noutput: [parents great_pret pretentious usual\nrecommend \nnot_recom 1440 1440 1440\npriority 858 1484 1924\nrecommend 0 0 2\nspec_prior 2022 1264 758\nvery_recom 0 132 196]\ntimestamp: 2025-03-06T19:39:46.980090\ncomments: []\nexception: \n\nuid: output_3\nstatus: fail\ntype: table\nproperties: {\'method\': \'crosstab\'}\nsdc: {\'summary\': {\'suppressed\': True, \'negative\': 0, \'missing\': 0, \'threshold\': 1, \'p-ratio\': 4, \'nk-rule\': 4, \'all-values-are-same\': 0}, \'cells\': {\'negative\': [], \'missing\': [], \'threshold\': [[2, 2]], \'p-ratio\': [[2, 0], [2, 1], [2, 2], [4, 0]], \'nk-rule\': [[2, 0], [2, 1], [2, 2], [4, 0]], \'all-values-are-same\': []}}\ncommand: safe_table = acro.crosstab(\nsummary: fail; threshold: 1 cells suppressed; p-ratio: 4 cells suppressed; nk-rule: 4 cells suppressed; \noutcome: parents great_pret pretentious \\\nrecommend \nnot_recom ok ok \npriority ok ok \nrecommend p-ratio; nk-rule; p-ratio; nk-rule; \nspec_prior ok ok \nvery_recom p-ratio; nk-rule; ok \n\nparents usual \nrecommend \nnot_recom ok \npriority ok \nrecommend threshold; p-ratio; nk-rule; \nspec_prior ok \nvery_recom ok \noutput: [parents great_pret pretentious usual\nrecommend \nnot_recom 1440.0 1440.0 1440.0\npriority 858.0 1484.0 1924.0\nrecommend NaN NaN NaN\nspec_prior 2022.0 1264.0 758.0\nvery_recom NaN 132.0 196.0]\ntimestamp: 2025-03-06T19:39:47.019919\ncomments: []\nexception: \n\nuid: output_4\nstatus: fail\ntype: table\nproperties: {\'method\': \'crosstab\'}\nsdc: {\'summary\': {\'suppressed\': True, \'negative\': 0, \'missing\': 0, \'threshold\': 2, \'p-ratio\': 8, \'nk-rule\': 8, \'all-values-are-same\': 0}, \'cells\': {\'negative\': [], \'missing\': [], \'threshold\': [[2, 2], [2, 5]], \'p-ratio\': [[2, 0], [2, 1], [2, 2], [2, 3], [2, 4], [2, 5], [4, 0], [4, 3]], \'nk-rule\': [[2, 0], [2, 1], [2, 2], [2, 3], [2, 4], [2, 5], [4, 0], [4, 3]], \'all-values-are-same\': []}}\ncommand: safe_table = acro.crosstab(\nsummary: fail; threshold: 2 cells suppressed; p-ratio: 8 cells suppressed; nk-rule: 8 cells suppressed; \noutcome: mode_aggfunc \\\nparents great_pret pretentious \nrecommend \nnot_recom ok ok \npriority ok ok \nrecommend p-ratio; nk-rule; p-ratio; nk-rule; \nspec_prior ok ok \nvery_recom p-ratio; nk-rule; ok \n\n mean \\\nparents usual great_pret \nrecommend \nnot_recom ok ok \npriority ok ok \nrecommend threshold; p-ratio; nk-rule; p-ratio; nk-rule; \nspec_prior ok ok \nvery_recom ok p-ratio; nk-rule; \n\n \nparents pretentious usual \nrecommend \nnot_recom ok ok \npriority ok ok \nrecommend p-ratio; nk-rule; threshold; p-ratio; nk-rule; \nspec_prior ok ok \nvery_recom ok ok \noutput: [ mode_aggfunc mean \nparents great_pret pretentious usual great_pret pretentious usual\nrecommend \nnot_recom 2.0 1.0 1.0 3.125694 3.105556 3.074306\npriority 1.0 1.0 1.0 2.665501 3.030323 3.116944\nrecommend NaN NaN NaN NaN NaN NaN\nspec_prior 3.0 3.0 3.0 3.353610 3.370253 3.393140\nvery_recom NaN 1.0 1.0 NaN 2.204545 2.244898]\ntimestamp: 2025-03-06T19:39:47.068066\ncomments: []\nexception: \n\nuid: output_5\nstatus: pass\ntype: table\nproperties: {\'method\': \'pivot_table\'}\nsdc: {\'summary\': {\'suppressed\': True, \'negative\': 0, \'missing\': 0, \'threshold\': 0, \'p-ratio\': 0, \'nk-rule\': 0, \'all-values-are-same\': 0}, \'cells\': {\'negative\': [], \'missing\': [], \'threshold\': [], \'p-ratio\': [], \'nk-rule\': [], \'all-values-are-same\': []}}\ncommand: table = acro.pivot_table(\nsummary: pass\noutcome: mean std\n children children\nparents \ngreat_pret ok ok\npretentious ok ok\nusual ok ok\noutput: [ mean std\n children children\nparents \ngreat_pret 3.140972 2.270396\npretentious 3.129630 2.250436\nusual 3.110648 2.213072]\ntimestamp: 2025-03-06T19:39:47.105651\ncomments: []\nexception: \n\nuid: output_6\nstatus: fail\ntype: table\nproperties: {\'method\': \'pivot_table\'}\nsdc: {\'summary\': {\'suppressed\': True, \'negative\': 0, \'missing\': 0, \'threshold\': 5, \'p-ratio\': 5, \'nk-rule\': 5, \'all-values-are-same\': 0}, \'cells\': {\'negative\': [], \'missing\': [], \'threshold\': [[0, 2], [0, 4], [1, 2], [2, 2], [3, 2]], \'p-ratio\': [[0, 2], [0, 4], [1, 2], [2, 2], [3, 2]], \'nk-rule\': [[0, 2], [0, 4], [1, 2], [2, 2], [3, 2]], \'all-values-are-same\': []}}\ncommand: safe_table = acro.pivot_table(\nsummary: fail; threshold: 5 cells suppressed; p-ratio: 5 cells suppressed; nk-rule: 5 cells suppressed; \noutcome: children \\\nrecommend not_recom priority recommend spec_prior \nparents \ngreat_pret ok ok threshold; p-ratio; nk-rule; ok \npretentious ok ok threshold; p-ratio; nk-rule; ok \nusual ok ok threshold; p-ratio; nk-rule; ok \nAll ok ok threshold; p-ratio; nk-rule; ok \n\n \nrecommend very_recom All \nparents \ngreat_pret threshold; p-ratio; nk-rule; ok \npretentious ok ok \nusual ok ok \nAll ok ok \noutput: [ children \nrecommend not_recom priority spec_prior very_recom All\nparents \ngreat_pret 3.125694 2.665501 3.353610 NaN 3.140972\npretentious 3.105556 3.030323 3.370253 2.204545 3.129630\nusual 3.074306 3.116944 3.393140 2.244898 3.111626\nAll 3.101852 2.996015 3.366222 2.228659 3.127412]\ntimestamp: 2025-03-06T19:39:47.231513\ncomments: []\nexception: \n\nuid: output_7\nstatus: pass\ntype: regression\nproperties: {\'method\': \'ols\', \'dof\': 12958.0}\nsdc: {}\ncommand: results = acro.ols(y, x)\nsummary: pass; dof=12958.0 >= 10\noutcome: Empty DataFrame\nColumns: []\nIndex: []\noutput: [ recommend R-squared: 0.001\nDep. Variable: \nModel: OLS Adj. R-squared: 0.001000\nMethod: Least Squares F-statistic: 13.830000\nDate: Thu, 06 Mar 2025 Prob (F-statistic): 0.000201\nTime: 19:39:47 Log-Likelihood: -25121.000000\nNo. Observations: 12960 AIC: 50250.000000\nDf Residuals: 12958 BIC: 50260.000000\nDf Model: 1 NaN NaN\nCovariance Type: nonrobust NaN NaN, coef std err t P>|t| [0.025 0.975]\nconst 2.2099 0.025 87.263 0.0 2.160 2.260\nchildren 0.0245 0.007 3.718 0.0 0.012 0.037, 77090.215 Durbin-Watson: 2.883\nOmnibus: \nProb(Omnibus): 0.000 Jarque-Bera (JB): 1741.57\nSkew: -0.486 Prob(JB): 0.00\nKurtosis: 1.489 Cond. No. 6.90]\ntimestamp: 2025-03-06T19:39:47.388052\ncomments: []\nexception: \n\nuid: output_8\nstatus: pass\ntype: regression\nproperties: {\'method\': \'olsr\', \'dof\': 12958.0}\nsdc: {}\ncommand: results = acro.olsr(formula="recommend ~ children", data=new_df)\nsummary: pass; dof=12958.0 >= 10\noutcome: Empty DataFrame\nColumns: []\nIndex: []\noutput: [ recommend R-squared: 0.001\nDep. Variable: \nModel: OLS Adj. R-squared: 0.001000\nMethod: Least Squares F-statistic: 13.830000\nDate: Thu, 06 Mar 2025 Prob (F-statistic): 0.000201\nTime: 19:39:47 Log-Likelihood: -25121.000000\nNo. Observations: 12960 AIC: 50250.000000\nDf Residuals: 12958 BIC: 50260.000000\nDf Model: 1 NaN NaN\nCovariance Type: nonrobust NaN NaN, coef std err t P>|t| [0.025 0.975]\nIntercept 2.2099 0.025 87.263 0.0 2.160 2.260\nchildren 0.0245 0.007 3.718 0.0 0.012 0.037, 77090.215 Durbin-Watson: 2.883\nOmnibus: \nProb(Omnibus): 0.000 Jarque-Bera (JB): 1741.57\nSkew: -0.486 Prob(JB): 0.00\nKurtosis: 1.489 Cond. No. 6.90]\ntimestamp: 2025-03-06T19:39:47.414293\ncomments: []\nexception: \n\nuid: output_9\nstatus: pass\ntype: regression\nproperties: {\'method\': \'probit\', \'dof\': 12958.0}\nsdc: {}\ncommand: results = acro.probit(y, x)\nsummary: pass; dof=12958.0 >= 10\noutcome: Empty DataFrame\nColumns: []\nIndex: []\noutput: [ finance No. Observations: 12960\nDep. Variable: \nModel: Probit Df Residuals: 12958.000000\nMethod: MLE Df Model: 1.000000\nDate: Thu, 06 Mar 2025 Pseudo R-squ.: 0.000004\nTime: 19:39:47 Log-Likelihood: -8983.200000\nconverged: True LL-Null: -8983.200000\nCovariance Type: nonrobust LLR p-value: 0.799200, coef std err z P>|z| [0.025 0.975]\nconst -0.0039 0.019 -0.207 0.836 -0.041 0.033\nchildren 0.0012 0.005 0.254 0.799 -0.008 0.011]\ntimestamp: 2025-03-06T19:39:47.439598\ncomments: []\nexception: \n\nuid: output_10\nstatus: pass\ntype: regression\nproperties: {\'method\': \'logit\', \'dof\': 12958.0}\nsdc: {}\ncommand: results = acro.logit(y, x)\nsummary: pass; dof=12958.0 >= 10\noutcome: Empty DataFrame\nColumns: []\nIndex: []\noutput: [ finance No. Observations: 12960\nDep. Variable: \nModel: Logit Df Residuals: 12958.000000\nMethod: MLE Df Model: 1.000000\nDate: Thu, 06 Mar 2025 Pseudo R-squ.: 0.000004\nTime: 19:39:47 Log-Likelihood: -8983.200000\nconverged: True LL-Null: -8983.200000\nCovariance Type: nonrobust LLR p-value: 0.799200, coef std err z P>|z| [0.025 0.975]\nconst -0.0062 0.030 -0.207 0.836 -0.065 0.053\nchildren 0.0020 0.008 0.254 0.799 -0.013 0.017]\ntimestamp: 2025-03-06T19:39:47.457696\ncomments: []\nexception: \n\nuid: output_11\nstatus: fail\ntype: table\nproperties: {\'method\': \'surv_func\'}\nsdc: {\'summary\': {\'suppressed\': True, \'negative\': 0, \'missing\': 0, \'threshold\': 76, \'p-ratio\': 0, \'nk-rule\': 0, \'all-values-are-same\': 0}, \'cells\': {\'negative\': [], \'missing\': [], \'threshold\': [[1, 0], [1, 1], [1, 2], [1, 3], [2, 0], [2, 1], [2, 2], [2, 3], [3, 0], [3, 1], [3, 2], [3, 3], [4, 0], [4, 1], [4, 2], [4, 3], [5, 0], [5, 1], [5, 2], [5, 3], [6, 0], [6, 1], [6, 2], [6, 3], [7, 0], [7, 1], [7, 2], [7, 3], [8, 0], [8, 1], [8, 2], [8, 3], [9, 0], [9, 1], [9, 2], [9, 3], [10, 0], [10, 1], [10, 2], [10, 3], [11, 0], [11, 1], [11, 2], [11, 3], [12, 0], [12, 1], [12, 2], [12, 3], [13, 0], [13, 1], [13, 2], [13, 3], [14, 0], [14, 1], [14, 2], [14, 3], [15, 0], [15, 1], [15, 2], [15, 3], [16, 0], [16, 1], [16, 2], [16, 3], [17, 0], [17, 1], [17, 2], [17, 3], [18, 0], [18, 1], [18, 2], [18, 3], [19, 0], [19, 1], [19, 2], [19, 3]], \'p-ratio\': [], \'nk-rule\': [], \'all-values-are-same\': []}}\ncommand: safe_table = acro.surv_func(data.futime, data.death, output="table")\nsummary: fail; threshold: 76 cells suppressed; \noutcome: Surv_prob Surv_prob_SE num_at_risk num_events\nTime \n51 ok ok ok ok\n69 threshold; threshold; threshold; threshold; \n85 threshold; threshold; threshold; threshold; \n91 threshold; threshold; threshold; threshold; \n115 threshold; threshold; threshold; threshold; \n372 threshold; threshold; threshold; threshold; \n667 threshold; threshold; threshold; threshold; \n874 threshold; threshold; threshold; threshold; \n1039 threshold; threshold; threshold; threshold; \n1046 threshold; threshold; threshold; threshold; \n1281 threshold; threshold; threshold; threshold; \n1286 threshold; threshold; threshold; threshold; \n1326 threshold; threshold; threshold; threshold; \n1355 threshold; threshold; threshold; threshold; \n1626 threshold; threshold; threshold; threshold; \n1903 threshold; threshold; threshold; threshold; \n1914 threshold; threshold; threshold; threshold; \n2776 threshold; threshold; threshold; threshold; \n2851 threshold; threshold; threshold; threshold; \n3309 threshold; threshold; threshold; threshold; \noutput: [ Surv prob Surv prob SE num at risk num events\nTime \n51 0.95 0.048734 20.0 1.0\n69 NaN NaN NaN NaN\n85 NaN NaN NaN NaN\n91 NaN NaN NaN NaN\n115 NaN NaN NaN NaN\n372 NaN NaN NaN NaN\n667 NaN NaN NaN NaN\n874 NaN NaN NaN NaN\n1039 NaN NaN NaN NaN\n1046 NaN NaN NaN NaN\n1281 NaN NaN NaN NaN\n1286 NaN NaN NaN NaN\n1326 NaN NaN NaN NaN\n1355 NaN NaN NaN NaN\n1626 NaN NaN NaN NaN\n1903 NaN NaN NaN NaN\n1914 NaN NaN NaN NaN\n2776 NaN NaN NaN NaN\n2851 NaN NaN NaN NaN\n3309 NaN NaN NaN NaN]\ntimestamp: 2025-03-06T19:39:48.298262\ncomments: []\nexception: \n\nuid: output_12\nstatus: fail\ntype: survival plot\nproperties: {\'method\': \'surv_func\'}\nsdc: {\'summary\': {\'suppressed\': True, \'negative\': 0, \'missing\': 0, \'threshold\': 76, \'p-ratio\': 0, \'nk-rule\': 0, \'all-values-are-same\': 0}, \'cells\': {\'negative\': [], \'missing\': [], \'threshold\': [[1, 0], [1, 1], [1, 2], [1, 3], [2, 0], [2, 1], [2, 2], [2, 3], [3, 0], [3, 1], [3, 2], [3, 3], [4, 0], [4, 1], [4, 2], [4, 3], [5, 0], [5, 1], [5, 2], [5, 3], [6, 0], [6, 1], [6, 2], [6, 3], [7, 0], [7, 1], [7, 2], [7, 3], [8, 0], [8, 1], [8, 2], [8, 3], [9, 0], [9, 1], [9, 2], [9, 3], [10, 0], [10, 1], [10, 2], [10, 3], [11, 0], [11, 1], [11, 2], [11, 3], [12, 0], [12, 1], [12, 2], [12, 3], [13, 0], [13, 1], [13, 2], [13, 3], [14, 0], [14, 1], [14, 2], [14, 3], [15, 0], [15, 1], [15, 2], [15, 3], [16, 0], [16, 1], [16, 2], [16, 3], [17, 0], [17, 1], [17, 2], [17, 3], [18, 0], [18, 1], [18, 2], [18, 3], [19, 0], [19, 1], [19, 2], [19, 3]], \'p-ratio\': [], \'nk-rule\': [], \'all-values-are-same\': []}}\ncommand: safe_plot = acro.surv_func(\nsummary: fail; threshold: 76 cells suppressed; \noutcome: Empty DataFrame\nColumns: []\nIndex: []\noutput: [\'acro_artifacts/kaplan-mier_0.png\']\ntimestamp: 2025-03-06T19:39:48.450221\ncomments: []\nexception: \n\n'
2: Remove some ACRO outputs before finalising#
Currently, all outputs names contain timestamp; that is the time when the output was created.
The output name can be taken from the outputs listed by the print_outputs function,
or by listing the results and choosing the specific output that needs to be removed
[26]:
acro.remove_output("output_0")
INFO:acro:records:remove(): output_0 removed
3: Rename ACRO outputs before finalising#
[27]:
acro.rename_output("output_2", "pivot_table")
INFO:acro:records:rename_output(): output_2 renamed to pivot_table
4: Add a comment to output#
[28]:
acro.add_comments("output_1", "Please let me have this data.")
acro.add_comments("output_1", "6 cells were suppressed in this table")
INFO:acro:records:a comment was added to output_1
INFO:acro:records:a comment was added to output_1
5: Add an unsupported output to the list of outputs#
This is an example to add an unsupported outputs (such as images) to the list of outputs
[29]:
acro.custom_output(
"XandY.jpeg", "This output is an image showing the relationship between X and Y"
)
INFO:acro:records:add_custom(): output_13
6: (the big one) Finalise ACRO#
This is an example of the function finalise() which the users must call at the end of each session.
It takes each output and saves it to a CSV file.
It also saves the SDC analysis for each output to a json file or Excel file (depending on the extension of the name of the file provided as an input to the function)
[30]:
output = acro.finalise("NURSERY", "json")
INFO:acro:records:
uid: output_1
status: fail
type: table
properties: {'method': 'crosstab'}
sdc: {'summary': {'suppressed': True, 'negative': 0, 'missing': 0, 'threshold': 5, 'p-ratio': 0, 'nk-rule': 0, 'all-values-are-same': 0}, 'cells': {'negative': [], 'missing': [], 'threshold': [[2, 0], [2, 1], [2, 2], [2, 3], [4, 0]], 'p-ratio': [], 'nk-rule': [], 'all-values-are-same': []}}
command: safe_table = acro.crosstab(df.recommend, df.parents, margins=True)
summary: fail; threshold: 5 cells suppressed;
outcome: parents great_pret pretentious usual All
recommend
not_recom ok ok ok ok
priority ok ok ok ok
recommend threshold; threshold; threshold; threshold;
spec_prior ok ok ok ok
very_recom threshold; ok ok ok
All ok ok ok ok
output: [parents great_pret pretentious usual All
recommend
not_recom 1440.0 1440 1440 4320
priority 858.0 1484 1924 4266
spec_prior 2022.0 1264 758 4044
very_recom NaN 132 196 328
All 4320.0 4320 4318 12958]
timestamp: 2025-03-06T19:39:46.961631
comments: ['Please let me have this data.', '6 cells were suppressed in this table']
exception:
The status of the record above is: fail.
Please explain why an exception should be granted.
suppressed
INFO:acro:records:
uid: output_3
status: fail
type: table
properties: {'method': 'crosstab'}
sdc: {'summary': {'suppressed': True, 'negative': 0, 'missing': 0, 'threshold': 1, 'p-ratio': 4, 'nk-rule': 4, 'all-values-are-same': 0}, 'cells': {'negative': [], 'missing': [], 'threshold': [[2, 2]], 'p-ratio': [[2, 0], [2, 1], [2, 2], [4, 0]], 'nk-rule': [[2, 0], [2, 1], [2, 2], [4, 0]], 'all-values-are-same': []}}
command: safe_table = acro.crosstab(
summary: fail; threshold: 1 cells suppressed; p-ratio: 4 cells suppressed; nk-rule: 4 cells suppressed;
outcome: parents great_pret pretentious \
recommend
not_recom ok ok
priority ok ok
recommend p-ratio; nk-rule; p-ratio; nk-rule;
spec_prior ok ok
very_recom p-ratio; nk-rule; ok
parents usual
recommend
not_recom ok
priority ok
recommend threshold; p-ratio; nk-rule;
spec_prior ok
very_recom ok
output: [parents great_pret pretentious usual
recommend
not_recom 1440.0 1440.0 1440.0
priority 858.0 1484.0 1924.0
recommend NaN NaN NaN
spec_prior 2022.0 1264.0 758.0
very_recom NaN 132.0 196.0]
timestamp: 2025-03-06T19:39:47.019919
comments: []
exception:
The status of the record above is: fail.
Please explain why an exception should be granted.
exception requested
INFO:acro:records:
uid: output_4
status: fail
type: table
properties: {'method': 'crosstab'}
sdc: {'summary': {'suppressed': True, 'negative': 0, 'missing': 0, 'threshold': 2, 'p-ratio': 8, 'nk-rule': 8, 'all-values-are-same': 0}, 'cells': {'negative': [], 'missing': [], 'threshold': [[2, 2], [2, 5]], 'p-ratio': [[2, 0], [2, 1], [2, 2], [2, 3], [2, 4], [2, 5], [4, 0], [4, 3]], 'nk-rule': [[2, 0], [2, 1], [2, 2], [2, 3], [2, 4], [2, 5], [4, 0], [4, 3]], 'all-values-are-same': []}}
command: safe_table = acro.crosstab(
summary: fail; threshold: 2 cells suppressed; p-ratio: 8 cells suppressed; nk-rule: 8 cells suppressed;
outcome: mode_aggfunc \
parents great_pret pretentious
recommend
not_recom ok ok
priority ok ok
recommend p-ratio; nk-rule; p-ratio; nk-rule;
spec_prior ok ok
very_recom p-ratio; nk-rule; ok
mean \
parents usual great_pret
recommend
not_recom ok ok
priority ok ok
recommend threshold; p-ratio; nk-rule; p-ratio; nk-rule;
spec_prior ok ok
very_recom ok p-ratio; nk-rule;
parents pretentious usual
recommend
not_recom ok ok
priority ok ok
recommend p-ratio; nk-rule; threshold; p-ratio; nk-rule;
spec_prior ok ok
very_recom ok ok
output: [ mode_aggfunc mean
parents great_pret pretentious usual great_pret pretentious usual
recommend
not_recom 2.0 1.0 1.0 3.125694 3.105556 3.074306
priority 1.0 1.0 1.0 2.665501 3.030323 3.116944
recommend NaN NaN NaN NaN NaN NaN
spec_prior 3.0 3.0 3.0 3.353610 3.370253 3.393140
very_recom NaN 1.0 1.0 NaN 2.204545 2.244898]
timestamp: 2025-03-06T19:39:47.068066
comments: []
exception:
The status of the record above is: fail.
Please explain why an exception should be granted.
exception requested
INFO:acro:records:
uid: output_6
status: fail
type: table
properties: {'method': 'pivot_table'}
sdc: {'summary': {'suppressed': True, 'negative': 0, 'missing': 0, 'threshold': 5, 'p-ratio': 5, 'nk-rule': 5, 'all-values-are-same': 0}, 'cells': {'negative': [], 'missing': [], 'threshold': [[0, 2], [0, 4], [1, 2], [2, 2], [3, 2]], 'p-ratio': [[0, 2], [0, 4], [1, 2], [2, 2], [3, 2]], 'nk-rule': [[0, 2], [0, 4], [1, 2], [2, 2], [3, 2]], 'all-values-are-same': []}}
command: safe_table = acro.pivot_table(
summary: fail; threshold: 5 cells suppressed; p-ratio: 5 cells suppressed; nk-rule: 5 cells suppressed;
outcome: children \
recommend not_recom priority recommend spec_prior
parents
great_pret ok ok threshold; p-ratio; nk-rule; ok
pretentious ok ok threshold; p-ratio; nk-rule; ok
usual ok ok threshold; p-ratio; nk-rule; ok
All ok ok threshold; p-ratio; nk-rule; ok
recommend very_recom All
parents
great_pret threshold; p-ratio; nk-rule; ok
pretentious ok ok
usual ok ok
All ok ok
output: [ children
recommend not_recom priority spec_prior very_recom All
parents
great_pret 3.125694 2.665501 3.353610 NaN 3.140972
pretentious 3.105556 3.030323 3.370253 2.204545 3.129630
usual 3.074306 3.116944 3.393140 2.244898 3.111626
All 3.101852 2.996015 3.366222 2.228659 3.127412]
timestamp: 2025-03-06T19:39:47.231513
comments: []
exception:
The status of the record above is: fail.
Please explain why an exception should be granted.
some reason
INFO:acro:records:
uid: output_11
status: fail
type: table
properties: {'method': 'surv_func'}
sdc: {'summary': {'suppressed': True, 'negative': 0, 'missing': 0, 'threshold': 76, 'p-ratio': 0, 'nk-rule': 0, 'all-values-are-same': 0}, 'cells': {'negative': [], 'missing': [], 'threshold': [[1, 0], [1, 1], [1, 2], [1, 3], [2, 0], [2, 1], [2, 2], [2, 3], [3, 0], [3, 1], [3, 2], [3, 3], [4, 0], [4, 1], [4, 2], [4, 3], [5, 0], [5, 1], [5, 2], [5, 3], [6, 0], [6, 1], [6, 2], [6, 3], [7, 0], [7, 1], [7, 2], [7, 3], [8, 0], [8, 1], [8, 2], [8, 3], [9, 0], [9, 1], [9, 2], [9, 3], [10, 0], [10, 1], [10, 2], [10, 3], [11, 0], [11, 1], [11, 2], [11, 3], [12, 0], [12, 1], [12, 2], [12, 3], [13, 0], [13, 1], [13, 2], [13, 3], [14, 0], [14, 1], [14, 2], [14, 3], [15, 0], [15, 1], [15, 2], [15, 3], [16, 0], [16, 1], [16, 2], [16, 3], [17, 0], [17, 1], [17, 2], [17, 3], [18, 0], [18, 1], [18, 2], [18, 3], [19, 0], [19, 1], [19, 2], [19, 3]], 'p-ratio': [], 'nk-rule': [], 'all-values-are-same': []}}
command: safe_table = acro.surv_func(data.futime, data.death, output="table")
summary: fail; threshold: 76 cells suppressed;
outcome: Surv_prob Surv_prob_SE num_at_risk num_events
Time
51 ok ok ok ok
69 threshold; threshold; threshold; threshold;
85 threshold; threshold; threshold; threshold;
91 threshold; threshold; threshold; threshold;
115 threshold; threshold; threshold; threshold;
372 threshold; threshold; threshold; threshold;
667 threshold; threshold; threshold; threshold;
874 threshold; threshold; threshold; threshold;
1039 threshold; threshold; threshold; threshold;
1046 threshold; threshold; threshold; threshold;
1281 threshold; threshold; threshold; threshold;
1286 threshold; threshold; threshold; threshold;
1326 threshold; threshold; threshold; threshold;
1355 threshold; threshold; threshold; threshold;
1626 threshold; threshold; threshold; threshold;
1903 threshold; threshold; threshold; threshold;
1914 threshold; threshold; threshold; threshold;
2776 threshold; threshold; threshold; threshold;
2851 threshold; threshold; threshold; threshold;
3309 threshold; threshold; threshold; threshold;
output: [ Surv prob Surv prob SE num at risk num events
Time
51 0.95 0.048734 20.0 1.0
69 NaN NaN NaN NaN
85 NaN NaN NaN NaN
91 NaN NaN NaN NaN
115 NaN NaN NaN NaN
372 NaN NaN NaN NaN
667 NaN NaN NaN NaN
874 NaN NaN NaN NaN
1039 NaN NaN NaN NaN
1046 NaN NaN NaN NaN
1281 NaN NaN NaN NaN
1286 NaN NaN NaN NaN
1326 NaN NaN NaN NaN
1355 NaN NaN NaN NaN
1626 NaN NaN NaN NaN
1903 NaN NaN NaN NaN
1914 NaN NaN NaN NaN
2776 NaN NaN NaN NaN
2851 NaN NaN NaN NaN
3309 NaN NaN NaN NaN]
timestamp: 2025-03-06T19:39:48.298262
comments: []
exception:
The status of the record above is: fail.
Please explain why an exception should be granted.
some other reason
INFO:acro:records:
uid: output_12
status: fail
type: survival plot
properties: {'method': 'surv_func'}
sdc: {'summary': {'suppressed': True, 'negative': 0, 'missing': 0, 'threshold': 76, 'p-ratio': 0, 'nk-rule': 0, 'all-values-are-same': 0}, 'cells': {'negative': [], 'missing': [], 'threshold': [[1, 0], [1, 1], [1, 2], [1, 3], [2, 0], [2, 1], [2, 2], [2, 3], [3, 0], [3, 1], [3, 2], [3, 3], [4, 0], [4, 1], [4, 2], [4, 3], [5, 0], [5, 1], [5, 2], [5, 3], [6, 0], [6, 1], [6, 2], [6, 3], [7, 0], [7, 1], [7, 2], [7, 3], [8, 0], [8, 1], [8, 2], [8, 3], [9, 0], [9, 1], [9, 2], [9, 3], [10, 0], [10, 1], [10, 2], [10, 3], [11, 0], [11, 1], [11, 2], [11, 3], [12, 0], [12, 1], [12, 2], [12, 3], [13, 0], [13, 1], [13, 2], [13, 3], [14, 0], [14, 1], [14, 2], [14, 3], [15, 0], [15, 1], [15, 2], [15, 3], [16, 0], [16, 1], [16, 2], [16, 3], [17, 0], [17, 1], [17, 2], [17, 3], [18, 0], [18, 1], [18, 2], [18, 3], [19, 0], [19, 1], [19, 2], [19, 3]], 'p-ratio': [], 'nk-rule': [], 'all-values-are-same': []}}
command: safe_plot = acro.surv_func(
summary: fail; threshold: 76 cells suppressed;
outcome: Empty DataFrame
Columns: []
Index: []
output: ['acro_artifacts/kaplan-mier_0.png']
timestamp: 2025-03-06T19:39:48.450221
comments: []
exception:
The status of the record above is: fail.
Please explain why an exception should be granted.
suppressed
INFO:acro:records:
uid: pivot_table
status: fail
type: table
properties: {'method': 'crosstab'}
sdc: {'summary': {'suppressed': False, 'negative': 0, 'missing': 0, 'threshold': 4, 'p-ratio': 0, 'nk-rule': 0, 'all-values-are-same': 0}, 'cells': {'negative': [], 'missing': [], 'threshold': [[2, 0], [2, 1], [2, 2], [4, 0]], 'p-ratio': [], 'nk-rule': [], 'all-values-are-same': []}}
command: safe_table = acro.crosstab(df.recommend, df.parents)
summary: fail; threshold: 4 cells may need suppressing;
outcome: parents great_pret pretentious usual
recommend
not_recom ok ok ok
priority ok ok ok
recommend threshold; threshold; threshold;
spec_prior ok ok ok
very_recom threshold; ok ok
output: [parents great_pret pretentious usual
recommend
not_recom 1440 1440 1440
priority 858 1484 1924
recommend 0 0 2
spec_prior 2022 1264 758
very_recom 0 132 196]
timestamp: 2025-03-06T19:39:46.980090
comments: []
exception:
The status of the record above is: fail.
Please explain why an exception should be granted.
a reason is provided
INFO:acro:records:
uid: output_13
status: review
type: custom
properties: {}
sdc: {}
command: custom
summary: review
outcome: Empty DataFrame
Columns: []
Index: []
output: ['XandY.jpeg']
timestamp: 2025-03-06T19:39:48.518030
comments: ['This output is an image showing the relationship between X and Y']
exception:
The status of the record above is: review.
Please explain why an exception should be granted.
image is not disclosive
INFO:acro:records:outputs written to: NURSERY
7: Add a directory of outputs to an acro object#
This is an example of adding a list of files (produced by the researcher without using ACRO) to an acro object and creates a results file for checking.
[31]:
import shutil
table = pd.crosstab(df.recommend, df.parents)
# save the output table to a file and add this file to a directory
src_path = "test_add_to_acro"
file_path = "crosstab.pkl"
dest_path = "SDC_results"
if not os.path.exists(src_path):
table.to_pickle(file_path)
os.mkdir(src_path)
shutil.move(file_path, src_path, copy_function=shutil.copytree)
# add the output to acro
add_to_acro(src_path, dest_path)
INFO:acro:version: 0.4.8
INFO:acro:config: {'safe_threshold': 10, 'safe_dof_threshold': 10, 'safe_nk_n': 2, 'safe_nk_k': 0.9, 'safe_pratio_p': 0.1, 'check_missing_values': False, 'survival_safe_threshold': 10, 'zeros_are_disclosive': True}
INFO:acro:automatic suppression: False
INFO:acro:records:add_custom(): output_0
INFO:acro:records:rename_output(): output_0 renamed to crosstab.pkl
INFO:acro:records:
uid: crosstab.pkl
status: review
type: custom
properties: {}
sdc: {}
command: custom
summary: review
outcome: Empty DataFrame
Columns: []
Index: []
output: ['test_add_to_acro/crosstab.pkl']
timestamp: 2025-03-06T19:41:22.128464
comments: ['']
exception:
The status of the record above is: review.
Please explain why an exception should be granted.
pickle file need some explanation
INFO:acro:records:outputs written to: SDC_results